Despite many promising medicines for treating COVID-19 being tested in clinical trials (1;2), there is no cure available yet in most parts of the world. Recently, Remdesivir by Gilead Sciences (U.S.) becomes the first approved COVID-19 treatment in Europe but comes with considerable drawbacks (3). A lot of research efforts therefore continue to focus on finding additional novel drugs. To effectively fight a pandemic, developing different drugs is necessary. Vaccines, such as antibody therapies, are powerful for preventing viral infections but cannot be used to treat or cure active infections. Therefore, chemical drugs for the treatment of acute infections are also sorely needed.
In general, it is important to have more than one available drug for the acute treatment of a disease. For example, some patients might be allergic to a certain drug, or the drug is not compatible with other medications that the patient is taking. Another complication can be that the pathogens (i.e. viruses or bacteria) evolve the ability to defeat and become resistant to the drug designed to kill them. If drug resistance arises in the patient, switching to a different drug for effective treatment becomes necessary.
The rise of COVID-19 has sparked immense efforts in scientists all around the world to find new “druggable” viral proteins. Proteins are molecules that perform many different tasks in cells and allow viruses—including coronaviruses—to reproduce. Because of this, proteins required by coronaviruses to reproduce are prime drug targets, as stopping them from working halts viral production. Moreover, researchers have identified dozens of “host factors” that are human proteins promoting viral reproduction, expanding the list of druggable protein targets (4).
Structural Biology Fuels the Design of Novel Drugs
The quest to find better drugs that bind to target proteins is strongly fueled by an area of science called structural biology, which determines the three-dimensional shape—the “structures”—of a specific protein. Drugs are commonly small molecules that bind to a protein and modify its action (5). A good drug fits the shape of the target molecule like a key fits a keyhole. Therefore, seeing the structure of a molecule allows scientists to repurpose previously approved drugs or to rationally design effective new drugs (6). Determining the structures of protein targets can be a challenging task that is generally time and labor-consuming. To avoid redundancy and minimize the costs, scientists typically share their solved protein structures in the public databases, such as the Protein Data Bank (PDB). As of August 2020, a total of more than three hundred coronavirus-related protein structures have been shared by scientists in the PDB (7).
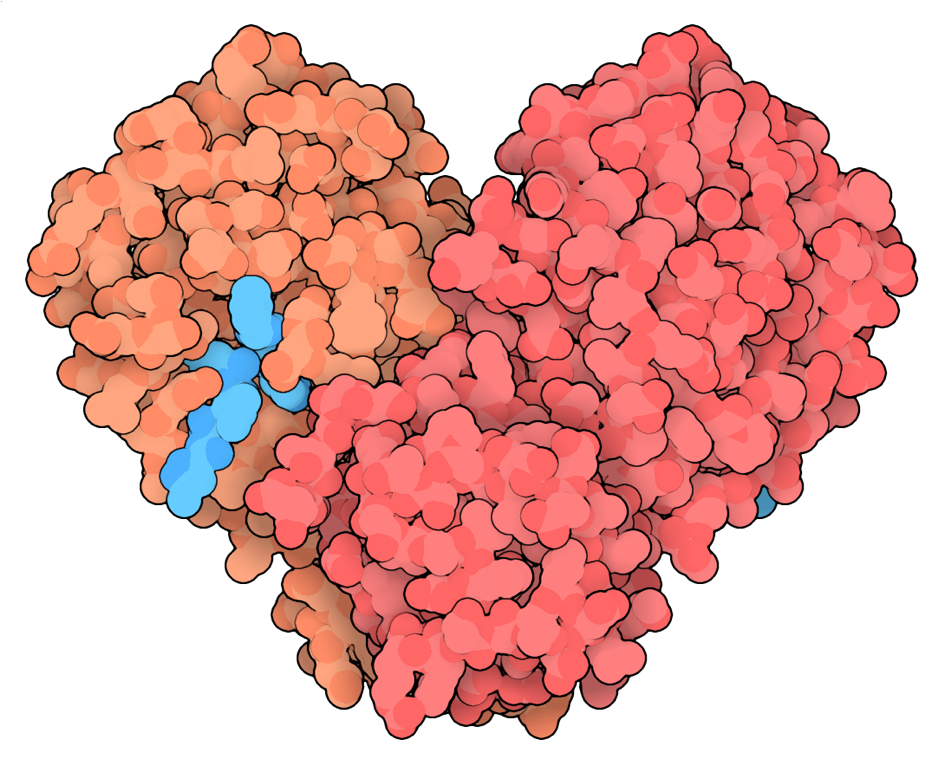
Of all these structures, nearly two hundred show a protein called Mpro, which is the main SARS-CoV-2 protease (a protein that breaks down other proteins or protein fragments) and is required by the virus to reproduce (8). These structures now help scientists design new drugs or improve already available drugs by allowing them to fine-tune the fit of drugs into the protease active site (9). These drugs—known as the “protease inhibitors”—stop the protease from working and halt the viral production. Many scientists now focus on Mpro because many similar proteases are long-known drug targets for treating viral infections with Human Immunodeficiency Virus (HIV) or Hepatitis C Virus (HCV) (10). Moreover, structures of Mpro have become available early on. This allowed many labs to quickly identify possible drugs that bind to the active site, which includes the FDA-approved disulfiram. Disulfiram is normally used to support alcoholism treatment (11) and is soon to be tested for the treatment of COVID-19 in the clinical trials (12).
In addition to Mpro, other viral and human proteins have been identified as important antiviral drug targets. The steadily increasing number of possible drug targets, the availability of their structures, and the worldwide collaborative research efforts are leading to the development of new drugs (13). Finding a cure for a disease can be a long and difficult path. However, the growing number of new drugs promises to pave the way and will certainly help to alleviate the severity of the disease and therefore save lives.
Sources and Further Reading
- Corum, Jonathan, Denise Grady, Sui-Lee Wee, and Carl Zimmer. “Coronavirus Vaccine Tracker.” The New York Times, June 10, 2020. https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html.
- “DrugBank.” https://www.drugbank.ca/.
- Kotsev, Victor. “Remdesivir Becomes the First Covid-19 Treatment Approved In Europe.” Eu, 8 July 2020, www.labiotech.eu/medical/covid-19-remdesivir-europe/?utm_source=newsletter&utm_medium=email&utm_campaign=campaign-294&mc_cid=1108c2d7bf&mc_eid=db0ba38dd2.
- Gordon, David E., Gwendolyn M. Jang, Mehdi Bouhaddou, Jiewei Xu, Kirsten Obernier, Kris M. White, Matthew J. O’Meara, et al. 2020. “A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing.” Nature 583 (7816): 459–68.
- Durrant, Jacob. “Lecture 1: How Drugs Work and What Makes a Good Drug Target.” March 16, 2017. https://durrantlab.pitt.edu/lecture-1-drugs-work/.
- “Structural Biology.” https://www.nigms.nih.gov/education/fact-sheets/Pages/structural-biology.aspx.
- “Rcsb Protein Data Bank.” https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true.
- “Coronavirus Proteases.” http://pdb101.rcsb.org/motm/242.
- Dai, Wenhao, Bing Zhang, Xia-Ming Jiang, Haixia Su, Jian Li, Yao Zhao, Xiong Xie, et al. 2020. “Structure-Based Design of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease.” Science 368 (6497): 1331–35.
- De Clercq, Erik, and Guangdi Li. 2016. “Approved Antiviral Drugs over the Past 50 Years.” Clinical Microbiology Reviews 29 (3): 695–747.
- Lin, Min-Han, David C. Moses, Chih-Hua Hsieh, Shu-Chun Cheng, Yau-Hung Chen, Chiao-Yin Sun, and Chi-Yuan Chou. 2018. “Disulfiram Can Inhibit MERS and SARS Coronavirus Papain-like Proteases via Different Modes.” Antiviral Research 150 (February): 155–63.
- “DISulfiram for COvid-19 (DISCO) Trial. ClinicalTrials.gov.” https://clinicaltrials.gov/ct2/show/NCT04485130.
- Fan, Shiyong, Dian Xiao, Yanming Wang, Lianqi Liu, Xinbo Zhou, and Wu Zhong. 2020. “Research Progress on Repositioning Drugs and Specific Therapeutic Drugs for SARS-CoV-2.” Future Medicinal Chemistry, July. https://doi.org/4155/fmc-2020-0158.
Image Source:
Structure of MPro, the SARS-CoV-2 (2019-nCoV) coronavirus main protease (in orange and red) Mpro, bound to a drug (in turquoise).
Image credit: David Goodsell, https://pdb101.rcsb.org/motm/242